Appendix: Support vector machines#
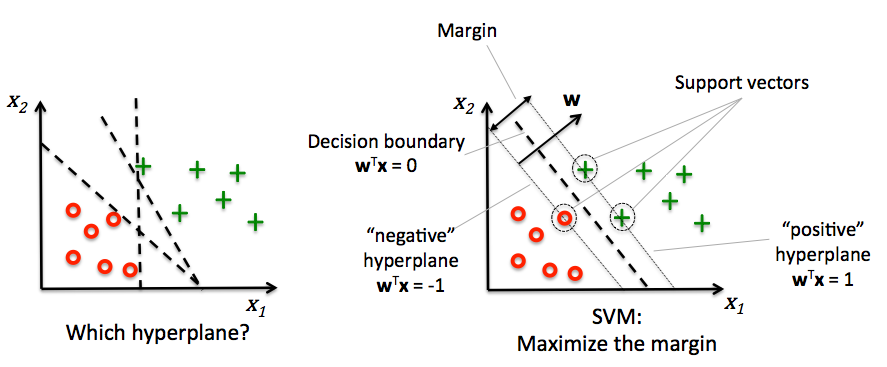
Decision boundaries close to training points may generalize badly
Very similar (nearby) test point are classified as the other class
Choose a boundary that is as far away from training points as possible
The support vectors are the training samples closest to the hyperplane
The margin is the distance between the separating hyperplane and the support vectors
Hence, our objective is to maximize the margin
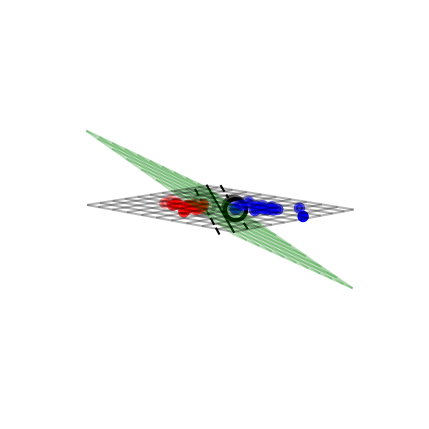
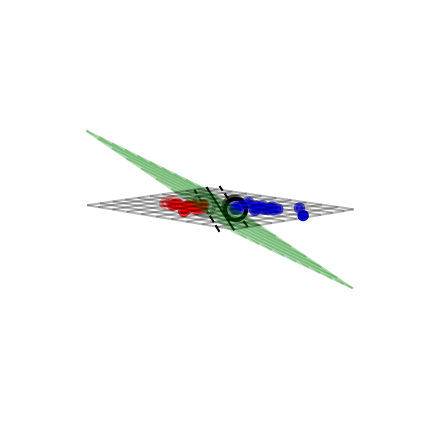
Solving SVMs with Lagrange Multipliers#
Imagine a hyperplane (green) \(y= \sum_1^p \mathbf{w}_i * \mathbf{x}_i + w_0\) that has slope \(\mathbf{w}\), value ‘+1’ for the positive (red) support vectors, and ‘-1’ for the negative (blue) ones
Margin between the boundary and support vectors is \(\frac{y-w_0}{||\mathbf{w}||}\), with \(||\mathbf{w}|| = \sum_i^p w_i^2\)
We want to find the weights that maximize \(\frac{1}{||\mathbf{w}||}\). We can also do that by maximizing \(\frac{1}{||\mathbf{w}||^2}\)
from sklearn.svm import SVC
# we create 40 separable points
np.random.seed(0)
sX = np.r_[np.random.randn(20, 2) - [2, 2], np.random.randn(20, 2) + [2, 2]]
sY = [0] * 20 + [1] * 20
# fit the model
s_clf = SVC(kernel='linear')
s_clf.fit(sX, sY)
@interact
def plot_svc_fit(rotationX=(0,20,1),rotationY=(90,180,1)):
# get the separating hyperplane
w = s_clf.coef_[0]
a = -w[0] / w[1]
xx = np.linspace(-5, 5)
yy = a * xx - (s_clf.intercept_[0]) / w[1]
zz = np.linspace(-2, 2, 30)
# plot the parallels to the separating hyperplane that pass through the
# support vectors
b = s_clf.support_vectors_[0]
yy_down = a * xx + (b[1] - a * b[0])
b = s_clf.support_vectors_[-1]
yy_up = a * xx + (b[1] - a * b[0])
# plot the line, the points, and the nearest vectors to the plane
fig = plt.figure(figsize=(7*fig_scale,4.5*fig_scale))
ax = plt.axes(projection="3d")
ax.plot3D(xx, yy, [0]*len(xx), 'k-')
ax.plot3D(xx, yy_down, [0]*len(xx), 'k--')
ax.plot3D(xx, yy_up, [0]*len(xx), 'k--')
ax.scatter3D(s_clf.support_vectors_[:, 0], s_clf.support_vectors_[:, 1], [0]*len(s_clf.support_vectors_[:, 0]),
s=85*fig_scale, edgecolors='k', c='w')
ax.scatter3D(sX[:, 0], sX[:, 1], [0]*len(sX[:, 0]), c=sY, cmap=plt.cm.bwr, s=10*fig_scale )
# Planes
XX, YY = np.meshgrid(xx, yy)
if interactive:
ZZ = w[0]*XX+w[1]*YY+clf.intercept_[0]
else: # rescaling (for prints) messes up the Z values
ZZ = w[0]*XX/fig_scale+w[1]*YY/fig_scale+clf.intercept_[0]*fig_scale/2
ax.plot_wireframe(XX, YY, XX*0, rstride=5, cstride=5, alpha=0.3, color='k', label='XY plane')
ax.plot_wireframe(XX, YY, ZZ, rstride=5, cstride=5, alpha=0.3, color='g', label='hyperplane')
ax.set_axis_off()
ax.view_init(rotationX, rotationY) # Use this to rotate the figure
ax.dist = 6
plt.tight_layout()
if not interactive:
plot_svc_fit(9,135)
Geometric interpretation#
We want to maximize \(f = \frac{1}{||w||^2}\) (blue contours)
The hyperplane (red) must be \(> 1\) for all positive examples:
\(g(\mathbf{w}) = \mathbf{w} \mathbf{x_i} + w_0 > 1 \,\,\, \forall{i}, y(i)=1\)Find the weights \(\mathbf{w}\) that satify \(g\) but maximize \(f\)
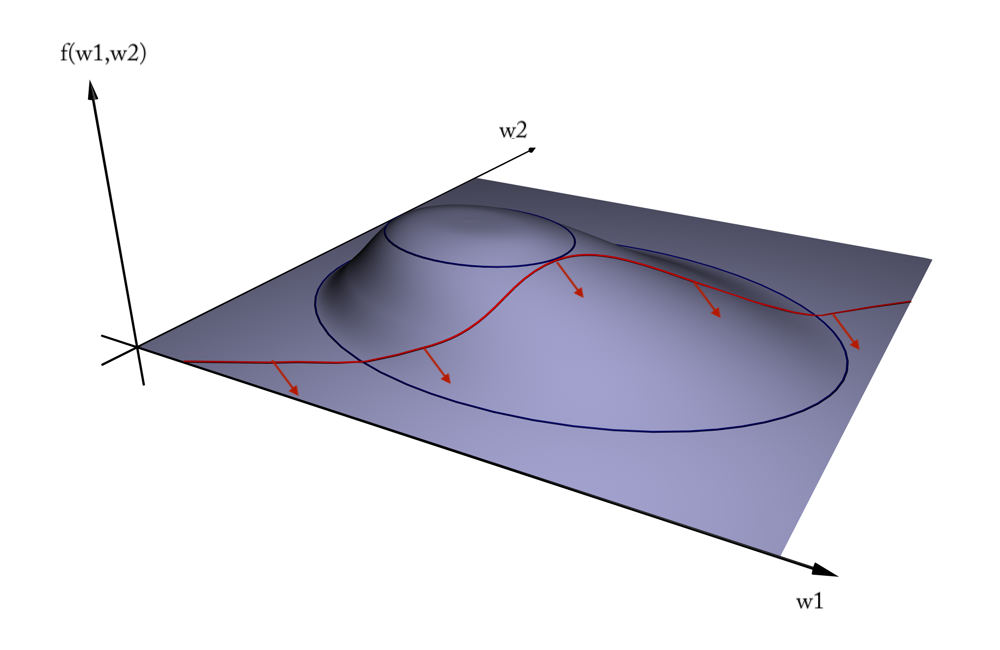
Solution#
A quadratic loss function with linear constraints can be solved with Lagrangian multipliers
This works by assigning a weight \(a_i\) (called a dual coefficient) to every data point \(x_i\)
They reflect how much individual points influence the weights \(\mathbf{w}\)
The points with non-zero \(a_i\) are the support vectors
Next, solve the following Primal objective:
\(y_i=\pm1\) is the correct class for example \(x_i\)
so that
It has a Dual formulation as well (See ‘Elements of Statistical Learning’ for the derivation):
so that
Computes the dual coefficients directly. A number \(l\) of these are non-zero (sparseness).
Dot product \(\mathbf{x_i} \mathbf{x_j}\) can be interpreted as the closeness between points \(\mathbf{x_i}\) and \(\mathbf{x_j}\)
\(\mathcal{L}_{Dual}\) increases if nearby support vectors \(\mathbf{x_i}\) with high weights \(a_i\) have different class \(y_i\)
\(\mathcal{L}_{Dual}\) also increases with the number of support vectors \(l\) and their weights \(a_i\)
Can be solved with quadratic programming, e.g. Sequential Minimal Optimization (SMO)
Example result. The circled samples are support vectors, together with their coefficients.
from sklearn.svm import SVC
# Plot SVM support vectors
def plot_linear_svm(X,y,C,ax):
clf = SVC(kernel='linear', C=C)
clf.fit(X, y)
# get the separating hyperplane
w = clf.coef_[0]
a = -w[0] / w[1]
xx = np.linspace(-5, 5)
yy = a * xx - (clf.intercept_[0]) / w[1]
# plot the parallels to the separating hyperplane
yy_down = (-1-w[0]*xx-clf.intercept_[0])/w[1]
yy_up = (1-w[0]*xx-clf.intercept_[0])/w[1]
# plot the line, the points, and the nearest vectors to the plane
ax.set_title('C = %s' % C)
ax.plot(xx, yy, 'k-')
ax.plot(xx, yy_down, 'k--')
ax.plot(xx, yy_up, 'k--')
ax.scatter(clf.support_vectors_[:, 0], clf.support_vectors_[:, 1],
s=85*fig_scale, edgecolors='gray', c='w', zorder=10, lw=1*fig_scale)
ax.scatter(X[:, 0], X[:, 1], c=y, zorder=10, cmap=plt.cm.bwr)
ax.axis('tight')
# Add coefficients
for i, coef in enumerate(clf.dual_coef_[0]):
ax.annotate("%0.2f" % (coef), (clf.support_vectors_[i, 0]+0.1,clf.support_vectors_[i, 1]+0.35), fontsize=10*fig_scale, zorder=11)
ax.set_xlim(np.min(X[:, 0])-0.5, np.max(X[:, 0])+0.5)
ax.set_ylim(np.min(X[:, 1])-0.5, np.max(X[:, 1])+0.5)
ax.set_xticks(())
ax.set_yticks(())
# we create 40 separable points
np.random.seed(0)
svm_X = np.r_[np.random.randn(20, 2) - [2, 2], np.random.randn(20, 2) + [2, 2]]
svm_Y = [0] * 20 + [1] * 20
svm_fig, svm_ax = plt.subplots(figsize=(8*fig_scale,5*fig_scale))
plot_linear_svm(svm_X,svm_Y,1,svm_ax)
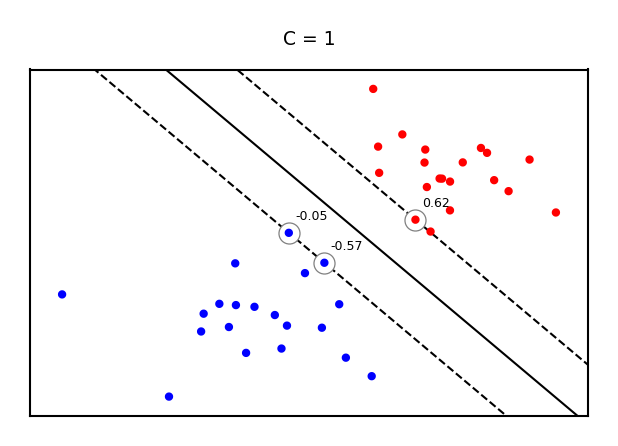
Making predictions#
\(a_i\) will be 0 if the training point lies on the right side of the decision boundary and outside the margin
The training samples for which \(a_i\) is not 0 are the support vectors
Hence, the SVM model is completely defined by the support vectors and their dual coefficients (weights)
Knowing the dual coefficients \(a_i\), we can find the weights \(w\) for the maximal margin separating hyperplane:
Hence, we can classify a new sample \(\mathbf{u}\) by looking at the sign of \(\mathbf{w}\mathbf{u}+w_0\)
SVMs and kNN#
Remember, we will classify a new point \(\mathbf{u}\) by looking at the sign of:
Weighted k-nearest neighbor is a generalization of the k-nearest neighbor classifier. It classifies points by evaluating:
Hence: SVM’s predict much the same way as k-NN, only:
They only consider the truly important points (the support vectors): much faster
The number of neighbors is the number of support vectors
The distance function is an inner product of the inputs
Regularized (soft margin) SVMs#
If the data is not linearly separable, (hard) margin maximization becomes meaningless
Relax the contraint by allowing an error \(\xi_{i}\): \(y_i (\mathbf{w}\mathbf{x_i} + w_0) \geq 1 - \xi_{i}\)
Or (since \(\xi_{i} \geq 0\)):
The sum over all points is called hinge loss: \(\sum_i^n \xi_{i}\)
Attenuating the error component with a hyperparameter \(C\), we get the objective
Can still be solved with quadratic programming
def hinge_loss(yHat, y):
if y == 1:
return np.maximum(0,1-yHat)
else:
return np.maximum(0,1+yHat)
fig, ax = plt.subplots(figsize=(6*fig_scale,2*fig_scale))
x = np.linspace(-2,2,100)
ax.plot(x,hinge_loss(x, 1),lw=2*fig_scale,c='b',label='true label = 1', linestyle='-')
ax.plot(x,hinge_loss(x, 0),lw=2*fig_scale,c='r',label='true label = 0', linestyle='-')
ax.set_xlabel(r"Prediction value $\hat{y}$")
ax.set_ylabel("Hinge loss")
plt.grid()
plt.legend()
<matplotlib.legend.Legend at 0x13691fca8f0>
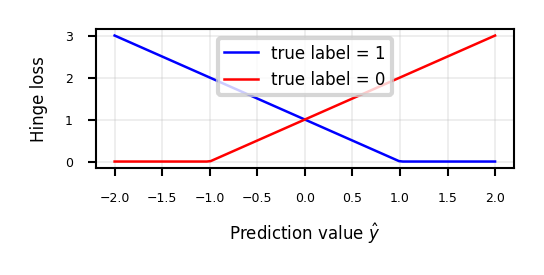
Least Squares SVMs#
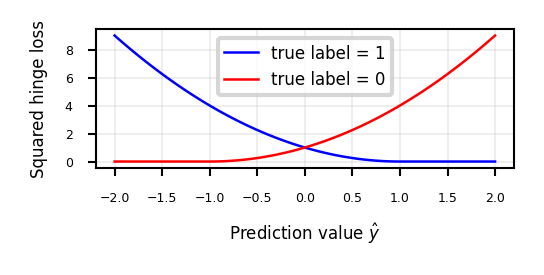
We can also use the squares of all the errors, or squared hinge loss: \(\sum_i^n \xi_{i}^2\)
This yields the Least Squares SVM objective
Can be solved with Lagrangian Multipliers and a set of linear equations
Still yields support vectors and still allows kernel trick
Support vectors are not sparse, but pruning techniques exist
fig, ax = plt.subplots(figsize=(6*fig_scale,2*fig_scale))
x = np.linspace(-2,2,100)
ax.plot(x,hinge_loss(x, 1)** 2,lw=2*fig_scale,c='b',label='true label = 1', linestyle='-')
ax.plot(x,hinge_loss(x, 0)** 2,lw=2*fig_scale,c='r',label='true label = 0', linestyle='-')
ax.set_xlabel(r"Prediction value $\hat{y}$")
ax.set_ylabel("Squared hinge loss")
plt.grid()
plt.legend();
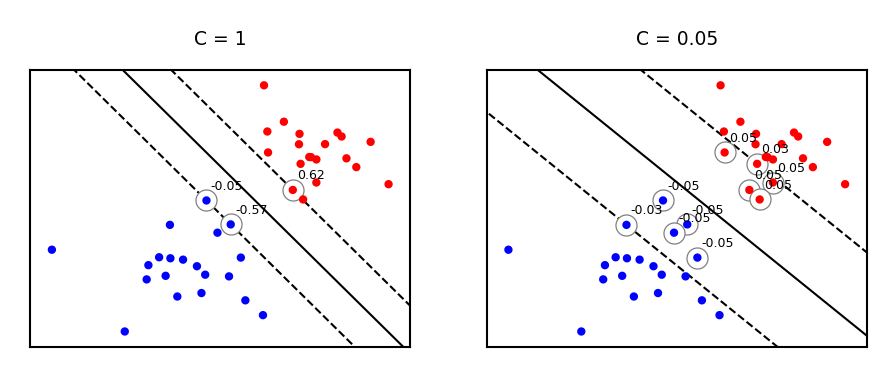
Effect of regularization on margin and support vectors#
SVM’s Hinge loss acts like L1 regularization, yields sparse models
C is the inverse regularization strength (inverse of \(\alpha\) in Lasso)
Larger C: fewer support vectors, smaller margin, more overfitting
Smaller C: more support vectors, wider margin, less overfitting
Needs to be tuned carefully to the data
fig, svm_axes = plt.subplots(nrows=1, ncols=2, figsize=(12*fig_scale, 4*fig_scale))
plot_linear_svm(svm_X,svm_Y,1,svm_axes[0])
plot_linear_svm(svm_X,svm_Y,0.05,svm_axes[1])
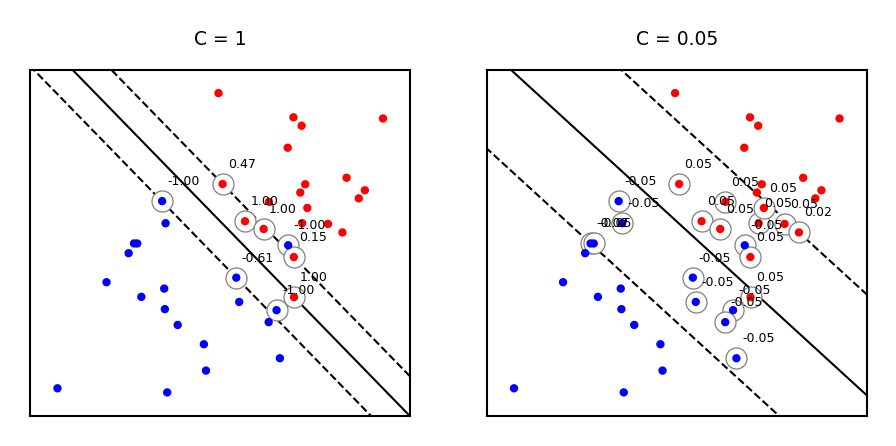
Same for non-linearly separable data
svm_X = np.r_[np.random.randn(20, 2) - [1, 1], np.random.randn(20, 2) + [1, 1]]
fig, svm_axes = plt.subplots(nrows=1, ncols=2, figsize=(12*fig_scale, 5*fig_scale))
plot_linear_svm(svm_X,svm_Y,1,svm_axes[0])
plot_linear_svm(svm_X,svm_Y,0.05,svm_axes[1])
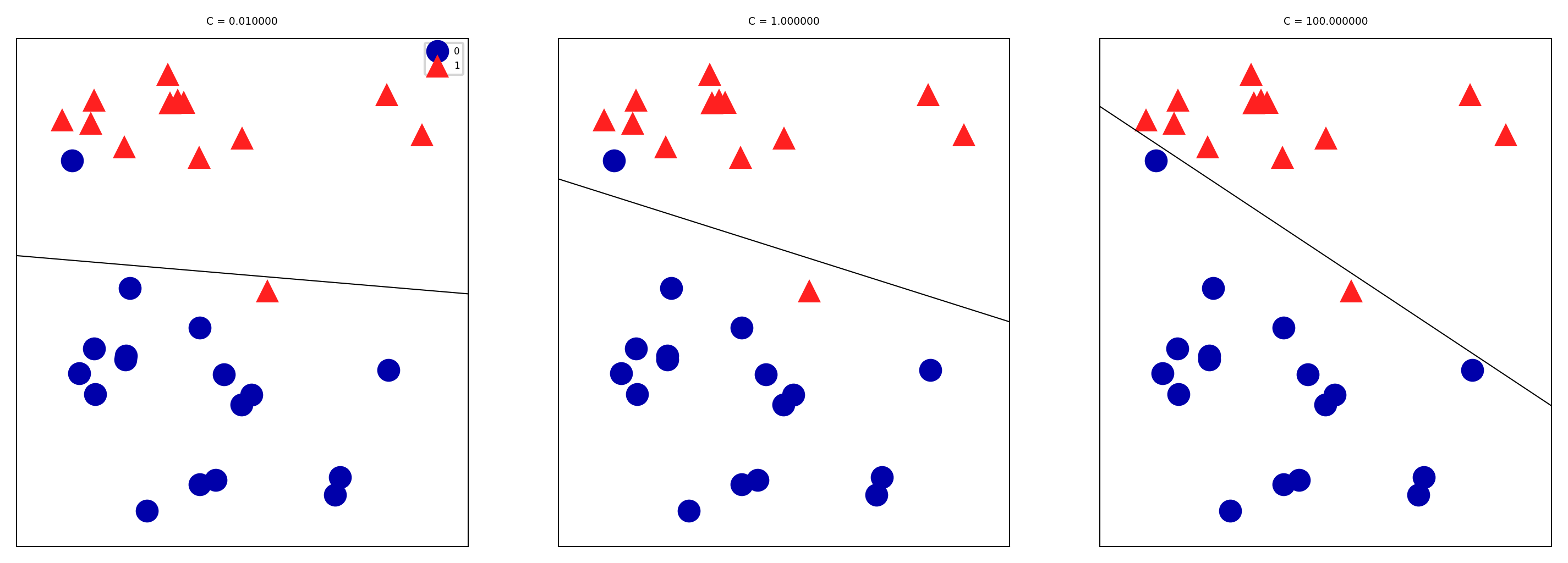
Large C values can lead to overfitting (e.g. fitting noise), small values can lead to underfitting
mglearn.plots.plot_linear_svc_regularization()
SVMs in scikit-learn#
svm.LinearSVC: faster for large datasetsAllows choosing between the primal or dual. Primal recommended when \(n\) >> \(p\)
Returns
coef_(\(\mathbf{w}\)) andintercept_(\(w_0\))
svm.SVCwithkernel=linear: allows kernel trick (see later)Also returns
support_vectors_(the support vectors) and thedual_coef_\(a_i\)Scales at least quadratically with the number of samples \(n\)
svm.LinearSVRandsvm.SVRare variants for regression
clf = svm.SVC(kernel='linear')
clf.fit(X, y)
print("Support vectors:", clf.support_vectors_[:])
print("Coefficients:", clf.dual_coef_[:])
from sklearn import svm
# Linearly separable dat
np.random.seed(0)
X = np.r_[np.random.randn(20, 2) - [2, 2], np.random.randn(20, 2) + [2, 2]]
y = [0] * 20 + [1] * 20
# Fit the model
clf = svm.SVC(kernel='linear')
clf.fit(X, y)
# Get the support vectors and weights
print("Support vectors:")
print(clf.support_vectors_[:])
print("Coefficients:")
print(clf.dual_coef_[:])
Support vectors:
[[-1.021 0.241]
[-0.467 -0.531]
[ 0.951 0.58 ]]
Coefficients:
[[-0.048 -0.569 0.617]]
Solving SVMs with Gradient Descent#
Soft-margin SVMs can, alternatively, be solved using gradient decent
Good for large datasets, but does not yield support vectors or kernel trick
Squared Hinge is differentiable
Hinge is not differentiable but convex, and has a subgradient:
Can be solved with (stochastic) gradient descent
fig, ax = plt.subplots(figsize=(6*fig_scale,2*fig_scale))
x = np.linspace(-2,2,100)
ax.plot(x,hinge_loss(x, 1),lw=2*fig_scale,c='b',label='true label = 1', linestyle='-')
ax.plot(x,hinge_loss(x, 0),lw=2*fig_scale,c='r',label='true label = 0', linestyle='-')
ax.set_xlabel(r"Prediction value $\hat{y}$")
ax.set_ylabel("Hinge loss")
plt.grid()
plt.legend()
<matplotlib.legend.Legend at 0x1369209a650>

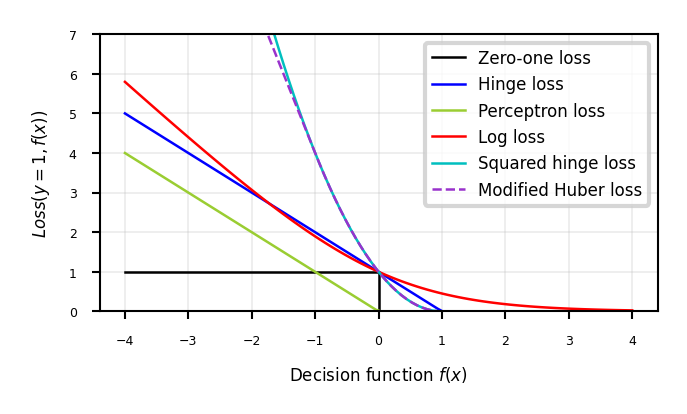
Generalized SVMs#
Because the derivative of hinge loss is undefined at y=1, smoothed versions are often used:
Squared hinge loss: yields least squares SVM
Equivalent to Ridge classification (with different solver)
Modified Huber loss: squared hinge, but linear after -1. Robust against outliers
Log loss can also be used (equivalent to logistic regression)
In sklearn,
SGDClassifiercan be used with any of these. Good for large datasets.
def modified_huber_loss(y_true, y_pred):
z = y_pred * y_true
loss = -4 * z
loss[z >= -1] = (1 - z[z >= -1]) ** 2
loss[z >= 1.] = 0
return loss
xmin, xmax = -4, 4
xx = np.linspace(xmin, xmax, 100)
lw = 2*fig_scale
fig, ax = plt.subplots(figsize=(8*fig_scale,4*fig_scale))
plt.plot([xmin, 0, 0, xmax], [1, 1, 0, 0], 'k-', lw=lw,
label="Zero-one loss")
plt.plot(xx, np.where(xx < 1, 1 - xx, 0), 'b-', lw=lw,
label="Hinge loss")
plt.plot(xx, -np.minimum(xx, 0), color='yellowgreen', lw=lw,
label="Perceptron loss")
plt.plot(xx, np.log2(1 + np.exp(-xx)), 'r-', lw=lw,
label="Log loss")
plt.plot(xx, np.where(xx < 1, 1 - xx, 0) ** 2, 'c-', lw=lw,
label="Squared hinge loss")
plt.plot(xx, modified_huber_loss(xx, 1), color='darkorchid', lw=lw,
linestyle='--', label="Modified Huber loss")
plt.ylim((0, 7))
plt.legend(loc="upper right")
plt.xlabel(r"Decision function $f(x)$")
plt.ylabel("$Loss(y=1, f(x))$")
plt.grid()
plt.legend()
<matplotlib.legend.Legend at 0x136922dd7b0>